Protein-Protein Interaction Networks
Co-immunoprecipitation (co-IP)
Co-IP in short, is a widely applied tool to identify physiologically-relevant protein-protein interactions by utilizing target protein-specific proteins derivatives like Antobodies and Enzyme to indirectly capture proteins that are bound to this specific target protein. As an extension of IP, Co-IP can capture and purify not only the primary target, but also other macromolecules binding to the target by native interactions. Difference between IP and co-IP is the focus of the experiment. IP is focused on the primary target, which binds the antibody. Whereas, Co-IP targets the secondary targets, which interacts with the primary proteins, instead of antibody. Following immunoprecipitation, proteins trapped on the beads are washed and eluted to gain purified primary and secondary target proteins. These target proteins can be characterized in various methods, such as WesternBlot and mass spec analysis under shotgun strategy, to identify partner protein IDs, binding affinities, kinetics of binding and undiscovered functions of the primary protein.
Critical factors about Co-IP:
As a powerful technique, Co-IP is utilized regularly by biochemists to explore protein–protein interactions. While the Co-IP methodology is straightforward, identifying physiological protein-protein interactions through Co-IP reaction is not easy, because of the instability of the interaction, nonspecific binding to IP reagents and antibody contamination. All of the problems above can have negative effects on detection of protein-protein interactions.
Co-IP of protein complexes
Since Co-IP depends on protein-protein interactions to detect the bound proteins, the binding affinity and stability of the physiological interactions during the whole Co-IP process is very important. Inadequate binding time and extensive washing steps may reduce the binding efficiency and cause a failure of detection of protein interaction. Therefore, for proteins, which are predicted to have week binding affinity or dynamic interactions, advance stabilization of protein-protein interactions are necessary, for instance, crosslinking processes can be performed before Co-IP.
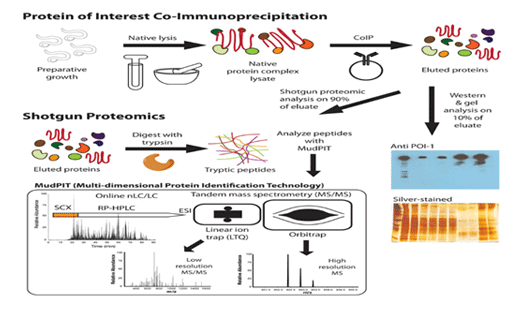
Importance of Co-IP service:
- Study the interactions between proteins and protein complex
- Monitor dynamics of protein interaction
- Study protein-protein interaction in a non-denaturing condition, which is almost physiological
- Mapping the interacting domains of proteins
Our Co-IP service contains:
- Experiment design based on your specific project needs: buffers, beads, antibodies, etc.
- Parameters optimization, such as binding time
- Cell lysis, IP, washing & elution
- WesternBlot/ mass spectrometry, depending on the project needs
- Data analysis & report delivery
Crosslinking Protein Interaction Analysis
Under natural Biological condition, most protein-protein interactions happen in a very short duration, which make them difficult to be studied. Crosslinking reagents, or crosslinkers, provide the analytical solution to capture protein-protein complexes by covalently binding them together as they interact, to freeze even transient, weak interactions for consequent isolation and characterization. The crosslinking can be performed in vivo or in vitro. Whether to use in vivo or in vitro crosslinking depends on the specific needs of the project. Proteins are crosslinked in a native state during In vivo crosslinking, whereas, for in vitro crosslinking protein may be denatured.
Importance of Crosslinking Protein Interaction Analysis
- Study impermanent in vivo protein-protein interaction
- Study low affinity in vitro protein-protein interactions
- Increase the detection sensitivity
Workflow of Crosslinking Protein Interaction Analysis
- Design the crosslinking method and type of crosslinking reagents based on your specific project needs: in vivo/ vitro
- Parameters optimization, such as binding and quenching time
- Cell lysis, CoIP
- WesternBlot/ mass spectrometry, depending on the project needs
- Data analysis & report delivery
Far-Western Blot Analysis
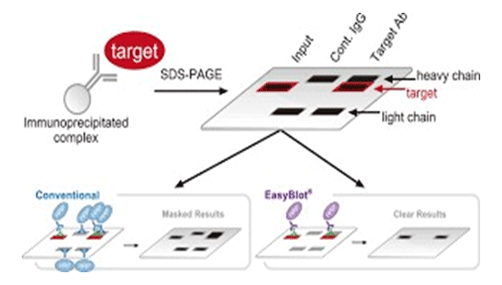
Far-Western blot has been widely used to study protein-protein interaction, cautions must be taken when preserving the native conformation and interaction conditions for the proteins. There are a few problems regarding far-Western blot. First, for proteins separated in a SDS gel, which is a denaturing condition, proteins are denatured. It is possible that denatured proteins may not interact with bait proteins, resulting in a failure to identify an interaction. Secondly, proteins are denatured and renature before binding assay, which may also leads to a potential conformational change of the protein, affecting the binding affinity of target proteins to the bait protein. Thirdly, if the exposure of binding motif is regulated by other factors, there can be a chance that the binding motif is buried inside and become unreachable to the bait, and cause a failure of detection of interaction. What’s more tricky is that, proteins presented in non-native conformations may interact in novel, artificial ways, resulting in false-positive interactions. Novelgene can design the experiment based on your needs, and assess the potential factors that may affect results. We promise reliable data with high reproducibility.
Importance of Far-Western Blot Analysis
- Study in vitro protein interactions
- Map protein binding domain
- Compare protein binding affinity
- No requirement for antibody
Workflow of Far-Western Blot Analysis
- Protein are quantified and separated by SDS or native PAGE
- Transfer proteins from gel to a membrane
- Transferred proteins are denatured and renatured to fully expose the binding motif
- The membrane is then blocked with BSA buffer
- The membrane is incubated with purified bait protein
- Signals of bait proteins are detected
- Images analysis & report delivery
Our Far-Western Blot methods contain:
- Direct detection of prey protein with a radioactive bait protein;
- Indirect detection with an antibody specific to the bait protein;
- Indirect detection with an antibody specific to the tag of a fusion-tagged bait protein;
- Indirect detection with a biotinylated bait protein and enzyme-conjugated avidin or streptavidin.
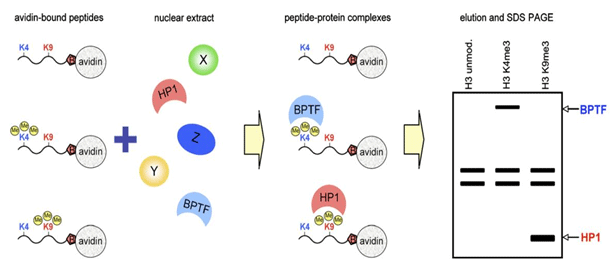
Pull-Down Assay
Pull-down assay is a simple and sensitive in vitro approach to studying protein-protein interactions (PPIs). In methodology, pull-down assay is similar to co-immunoprecipitation. In experiments, they both use an affinity ligand to capture interacting proteins. However, there is a difference between co-immunoprecipitation and the pull-down assay. The co-immunoprecipitation captures protein complexes by using immobilized antibodies, while the pull-down assay binds interacting proteins by using a purified and tagged protein, as “bait”.
Applications
- Study the direct interactions between proteins and protein complex.
- Verify predicted protein-protein interaction.
- Analyze and quantify the specific isoforms of or active/inactive protein in the tested samples.