Human Exome Sequencing
Researches further demonstrate that there are only 30 million base pairs of genes that contain essential information of proteins for human beings. The exome is usually defined as the sequence encompassing all exons of protein coding genes, as well as nonprotein coding elements such as microRNA or lncRNA. The investigation of exome helps to figure out which loci are responsible for proper diseases. When researchers plan to explore exons information of human genome, the cost to whole genome sequencing will be quite surprising considering the total length of human genome is over 3 billion base pairs in size. To study rare mendelian diseases, exome sequencing is a more effective way to identify the genetic variants. The breakthrough of target-enrichment strategies and DNA sequencing techniques contributes to the development of whole exome sequencing.
General workflow of exome sequencing:
Here a common workflow of exome sequencing is shown as below. The instructions of the major processes in the workflow will be discussed below.
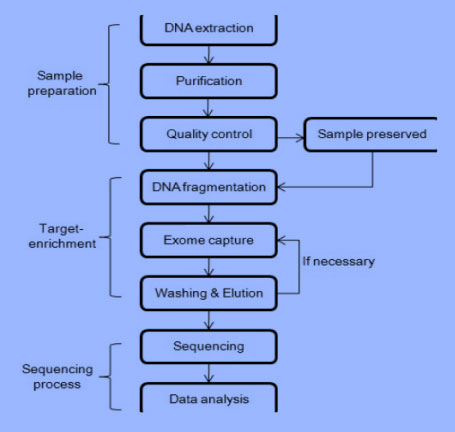
Prepare your DNA samples – DNA fragmentation: Almost all the experiment on DNA begins with DNA fragmentation. DNA should be sheared into proper pieces, because usually, the length of DNA sample extracted from tissues or cells is too long. This shearing process is called DNA fragmentation. Effective target length is determined by the sequencing instrument that you choose. In order to process whole exome sequencing, there are several major ways to fragmentize DNA samples.
Physical fragmentation. Physical fragmentation includes acoustic shearing, sonication and hydrodynamic shear. Among them acoustic shearing and sonication are the main methods for DNA fragmentation. DNA samples are broken into several pieces due to the acoustic cavitation and hydrodynamic shearing when they are exposed to ultrasound.
Enzymatic Methods. Enzymes used to break DNA into small pieces include nuclease or transposase. Nuclease will cleave the phosphodiester bonds between nucleic acids, resulting in the breaking down of DNA. Specifically, restriction endonuclease will cleave DNA at restriction sites. Transposase is used to mediate transposition events, processes that a certain DNA segment could “move around” the chromosome. It also plays a role in DNA fragmentation if we prepare appropriate DNA samples and transposase. The fragmentized DNA is linked with adapters instead of inserting again, resulting in fragmentation. After fragmentation, your DNA samples are ready for target-enrichment process.
Isolation of exome – target-enrichment methods: Exome has to be isolated from human genome before sequencing as the former contributes to only 1% of the latter. The process of capturing the target genomic regions is called target-enrichment. The basic idea of target-enrichment is to separate anything of interest from other substances using the physicochemical property difference between them. There are some common kits of target-enrichment methods. No matter what kit you choose, the variability in capture influences your exome sequencing, so be aware of the quality, quantity and fragment sizes of your DNA samples.


